ℹ️ Please Note
You are welcome to explore the available resources and video recordings. Please note that the resources in this course come from the Virtual Training 21 held in October 2025, and certificates are only issued to participants who attended the live webinar sessions.
Course description
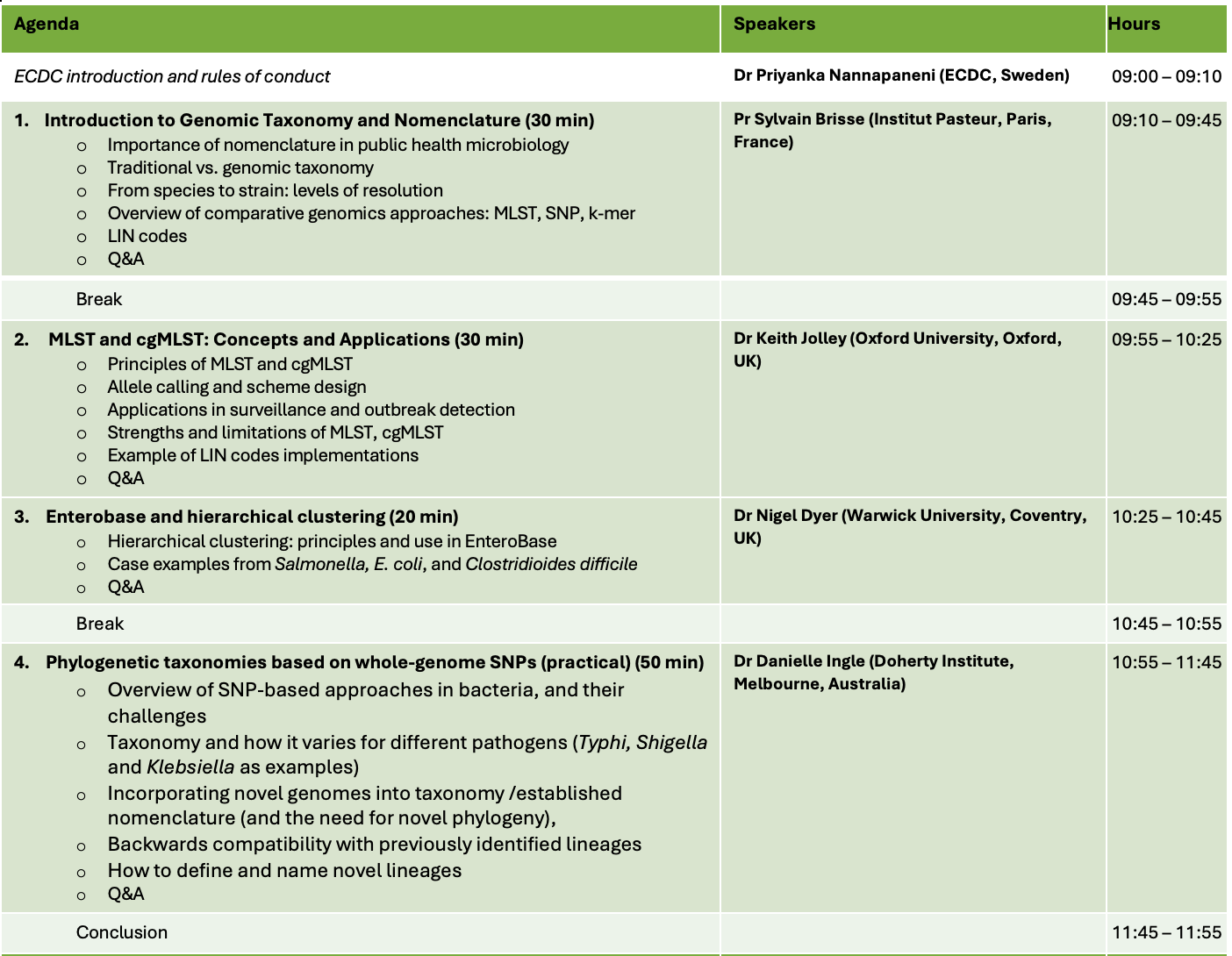
The aim of this virtual training workshop is to provide participants with a comprehensive understanding of bacterial classification and nomenclature approaches used in genomic surveillance. Participants will gain insight into MLST, cgMLST, SNP-based taxonomies, LIN codes, and k-mer-based clustering. The training will focus on how these systems are applied in public health microbiology to classify bacterial pathogens, track outbreaks, and support global surveillance efforts. Participants will also gain practical experience with tools such as BIGSdb, EnteroBase, MyKrobe, and PopPUNK, enabling them to apply bacterial taxonomy tools in genomic surveillance work..
Target audience
Public health microbiologists, epidemiologists, or bioinformaticians interested in learning about Bacterial strain Taxonomy for Genomic Surveillance.
Pre-requisites
For the practical exercises, participants may choose to use web-based (basic) tools or command-line (advanced) tools depending on their comfort and experience. Please see ‘Software requirements’ section below for more information.
Format
This virtual training consists of two half-day sessions held in the morning periods on Wednesday, October 22, and Thursday, October 23, from 09:00 to 12:00 (CEST).
You are visiting this course page as a guest user.😎
In case you enrolled for the live event, please login so you can access the discussion forum, provide feedback and download your certificate of participation.👍